Omnipose in 3D#
You can use the dim (dimension) argument to tell Omnipose to segment your images using a 3D model. This means that an image stack or 3D array is treated as a 3D volume given to a network trained on 3D volumes. This is very different from do_3D in Cellpose, which cleverly leveraged 2D predictions on all 2D slices of a 3D volume to construct a 3D flow field for segmentation. It turns out that the pseudo-ND Cellpose flows are an approximation to the true 3D flows of Omnipose, because the flows in each slice point to a local center of the cell, a.k.a. the cell skeleton to which the Omnipose field points. Thus, is not recommended to use Omnipose 2D slice predictions with do_3D. Instead, this notebook assumes you have trained a 3D model such as the plant_omni model.
1# Import dependencies
2import numpy as np
3from cellpose_omni import models, core
4
5# This checks to see if you have set up your GPU properly.
6# CPU performance is a lot slower, but not a problem if you
7# are only processing a few images.
8use_GPU = core.use_gpu()
9print('>>> GPU activated? %d'%use_GPU)
10
11# for plotting
12import matplotlib as mpl
13import matplotlib.pyplot as plt
14mpl.rcParams['figure.dpi'] = 300
15plt.style.use('dark_background')
16%matplotlib inline
2023-08-08 00:57:34,560 [INFO] ** TORCH GPU version installed and working. **
>>> GPU activated? 1
Read in data#
Here I am choosing one of the scaled-down volumes of the plant Arabidopsis thaliana dataset we used in the Omnipose paper.
1from pathlib import Path
2import os
3from cellpose_omni import io
4
5basedir = os.path.join(Path.cwd().parent,'test_files_3D')
6files = io.get_image_files(basedir)
7files # this displays the variable if it the last thing in the code block
['/home/kcutler/DataDrive/omnipose/docs/test_files_3D/Movie1_t00004_crop_gt.tif']
1from cellpose_omni import io, transforms
2from omnipose.utils import normalize99
3
4imgs = [io.imread(f) for f in files]
5
6# print some info about the images.
7for i in imgs:
8 print('Original image shape:',i.shape)
9 print('data type:',i.dtype)
10 print('data range:', i.min(),i.max())
11nimg = len(imgs)
12print('number of images:',nimg)
Original image shape: (162, 207, 443)
data type: uint8
data range: 0 247
number of images: 1
Initialize model#
plant_omni is the model trained on these plant cell images. (The image we loaded is from the test set, of course.)
1from cellpose_omni import models
2model_name = 'plant_omni'
3
4dim = 3
5nclasses = 3 # flow + dist + boundary
6nchan = 1
7omni = 1
8rescale = False
9diam_mean = 0
10use_GPU = 0 # Most people do not have enough VRAM to run on GPU... 24GB not enough for this image, need nearly 48GB
11model = models.CellposeModel(gpu=use_GPU, model_type=model_name, net_avg=False,
12 diam_mean=diam_mean, nclasses=nclasses, dim=dim, nchan=nchan)
2023-08-08 00:57:44,364 [INFO] >>plant_omni<< model set to be used
sdggsfgs
2023-08-08 00:57:44,364 [INFO] >>>> using CPU
Run segmentation#
1import torch
2torch.cuda.empty_cache()
3mask_threshold = -5 #usually this is -1
4flow_threshold = 0.
5diam_threshold = 12
6net_avg = False
7cluster = False
8verbose = 1
9tile = True
10chans = None
11compute_masks = 1
12resample=False
13rescale=None
14omni=True
15flow_factor = 10 # multiple to increase flow magnitude, useful in 3D
16transparency = True
17
18nimg = len(imgs)
19masks_om, flows_om = [[]]*nimg,[[]]*nimg
20
21# splitting the images into batches helps manage VRAM use so that memory can get properly released
22# here we have just one image, but most people will have several to process
23for k in range(nimg):
24 masks_om[k], flows_om[k], _ = model.eval(imgs[k],
25 channels=chans,
26 rescale=rescale,
27 mask_threshold=mask_threshold,
28 net_avg=net_avg,
29 transparency=transparency,
30 flow_threshold=flow_threshold,
31 omni=omni,
32 resample=resample,
33 verbose=verbose,
34 diam_threshold=diam_threshold,
35 cluster=cluster,
36 tile=tile,
37 compute_masks=compute_masks,
38 flow_factor=flow_factor)
2023-08-08 00:57:45,752 [INFO] Evaluating with flow_threshold 0.00, mask_threshold -5.00
2023-08-08 00:57:45,753 [INFO] using omni model, cluster False
2023-08-08 00:57:45,753 [INFO] not using dataparallel
2023-08-08 00:57:45,878 [INFO] multi-stack tiff read in as having 162 planes 1 channels
2023-08-08 00:58:36,584 [INFO] mask_threshold is -5.000000
2023-08-08 00:58:36,585 [INFO] Using hysteresis threshold.
dP_ times 10 for >2d, still experimenting
2023-08-08 00:58:37,615 [INFO] niter is None
2023-08-08 00:59:54,186 [INFO] Mean diameter is 25.683111
2023-08-08 00:59:54,307 [INFO] cluster: False, SKLEARN_ENABLED: True
2023-08-08 00:59:54,571 [INFO] nclasses: 5, mask.ndim: 3
2023-08-08 00:59:54,581 [INFO] Using boundary output to split edge defects.
2023-08-08 00:59:54,776 [INFO] Done finding masks.
2023-08-08 00:59:55,705 [INFO] compute_masks() execution time: 79.1 sec
2023-08-08 00:59:55,705 [INFO] execution time per pixel: 5.18728e-06 sec/px
2023-08-08 00:59:55,709 [INFO] execution time per cell pixel: 1.45631e-05 sec/px
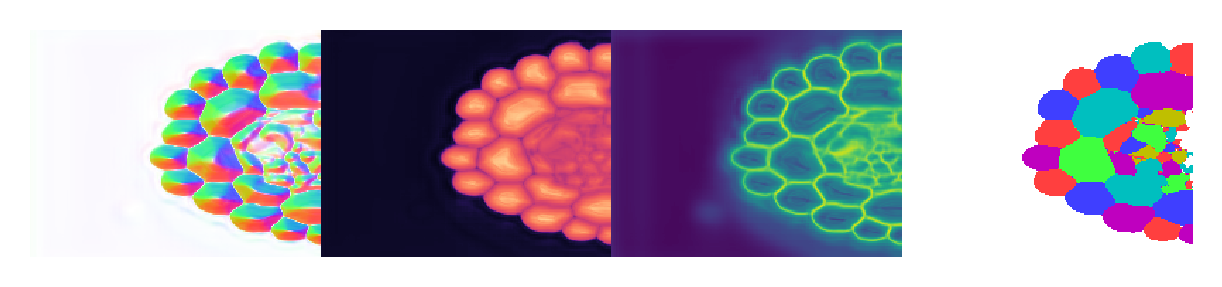
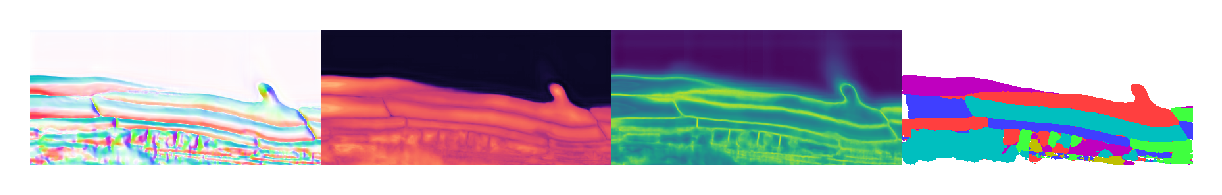
Plot results#
3D segmentation is a lot harder to show than 2D. If anyone figures out a good way to use one of the many tools out there (ipyvolume, K3D-Jupyter, itkwidgets, ipygany) for label visualization (not image volumes), please let me know. Few of these are in active development, and my own 3D work requires robust label editing tools anyway, which I do not think any available tools offer. Hence I shall just load in Napari and show you an auto-captured screenshot.
1%%capture
2import ncolor
3mask = masks_om[0]
4mask_nc = ncolor.label(mask,max_depth=20)
5
6import napari
7viewer = napari.view_labels(mask_nc);
8viewer.dims.ndisplay = 3
9viewer.camera.center = [s//2 for s in mask.shape]
10viewer.camera.zoom=1
11viewer.camera.angles=(10.90517458968619, -20.777067798396835, 58.04311170773853)
12viewer.camera.perspective=0.0
13viewer.camera.interactive=True
14
15img = viewer.screenshot(size=(1000,1000),scale=1,canvas_only=True,flash=False)
2023-08-08 00:59:58,954 [WARNING] Could not connect "org.freedesktop.IBus" to globalEngineChanged(QString)
1plt.figure(figsize=(3,3),frameon=False)
2plt.imshow(img)
3plt.axis('off')
4plt.show()

Plot orthogonal slices#
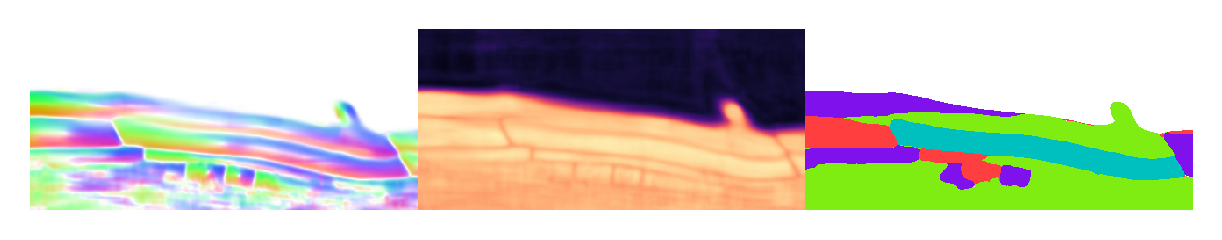
1from cellpose_omni import plot
2from omnipose.plot import apply_ncolor
3
4mu = flows_om[0][1]
5T = flows_om[0][2]
6bd = flows_om[0][4]
7# mu.shape,T.shape,bd.shape
8
9d = mu.shape[0]
10
11from omnipose.utils import rescale
12c = np.array([1]*2+[0]*(d-2))
13# c = np.arange(d)
14def cyclic_perm(a):
15 n = len(a)
16 b = [[a[i - j] for i in range(n)] for j in range(n)]
17 return b
18slices = []
19idx = np.arange(d)
20cmap = mpl.colormaps['magma']
21cmap2 = mpl.colormaps['viridis']
22
23for inds in cyclic_perm(c):
24 slc = tuple([slice(-1) if i else mu.shape[k+1]//2 for i,k in zip(inds,idx)])
25 flow = plot.dx_to_circ(mu[np.where(inds)+slc],transparency=1)/255
26 dist = cmap(rescale(T)[slc])
27 bnds = cmap2(rescale(bd)[slc])
28 msks = apply_ncolor(masks_om[0][slc])
29
30 fig = plt.figure(figsize=[5]*2,frameon=False)
31 plt.imshow(np.hstack((flow,dist,bnds,msks)),interpolation='none')
32 plt.axis('off')
33 plt.show()
34
4 -color algorthm failed,trying again with 5 colors. Depth 0
5 -color algorthm failed,trying again with 6 colors. Depth 1
4 -color algorthm failed,trying again with 5 colors. Depth 0
5 -color algorthm failed,trying again with 6 colors. Depth 1
Notes on the above#
Slices do not always look crisp because we are cutting though boundaries. At these locations, the flow and distance fields darken and the boundary field brightens. This can result in flat and muddled regions that are hard to interpret. Again, interactive 3D visualization tools are needed to properly evaluate the results of the segmentation. In this case, we have cut through the middle of enough cells to confirm that the output looks reasonable.
This small dataset with problematic annotations was sufficient for demonstrating that Omnipose can be used on 3D data, but I again emphasize that any algorithm will only work well after training on well-annotated, representative examples. In this case, small cell clusters were neither well-annotated nor well-represented in the training set, and you can see the negative impact of that in this example.
These 3D models are incredibly VRAM-hungry, so all results in the paper were actually run on an AWS instance. Here I ran them on CPU, which is much slower but necessary to do even with a 24GB Titan RTX.
Runing Omnipose with do_3D#
do_3D is not something you want to use with any Omnipose model, but you might want to use it with a 2D Cellpose model for 3D cells with extended shapes. This is because do_3D computes 2D flow fields from every yx, yz, and xz slice of the image and composites these components into a 3D field. It turns out that the center-seeking flow slices of Cellpose end up pointing roughly toward the local 3D skeleton, i.e. the do_3D Cellpose composite field approximates the true 3D flows of Omnipose. The 2D Omnipose field, on the other hand, cannot be composited into a useful 3D field.
Althought the do_3D Cellpose field directs pixels toward the skeleton, the stock Cellpose mask reconstruction algorithm tends to oversegment pixels into clusters along the skeleton. To avoid this, you can use a Cellpose model but with Omnipose mask reconstruciton by usin omni=True. Here is how to do this.
1from cellpose_omni import models, core
2
3# define cellpose model
4model_name = 'plant_cp'
5
6# this model was trained on 2D slices
7dim = 2
8nclasses = 2 # cellpose models have no boundary field, just flow and distance
9
10# Cellpose defaults to 2 channels;
11# this is the setup for grayscale in that case
12nchan = 2
13chans = [0,0]
14
15# no rescaling for this model
16diam_mean = 0
17
18
19use_GPU = core.use_gpu()
20model = models.CellposeModel(gpu=use_GPU, model_type=model_name, net_avg=False,
21 diam_mean=diam_mean, nclasses=nclasses, dim=dim, nchan=nchan)
22
23
24# segmentation parameters
25omni = 1
26rescale = False
27mask_threshold = 0
28net_avg = 0
29verbose = 0
30tile = 0
31compute_masks = 1
32rescale = None
33flow_threshold=0.
34do_3D=True
35flow_factor=10
36
37masks_cp, flows_cp, _ = model.eval(imgs,
38 channels=chans,
39 rescale=rescale,
40 mask_threshold=mask_threshold,
41 net_avg=net_avg,
42 transparency=True,
43 flow_threshold=flow_threshold,
44 verbose=verbose,
45 tile=tile,
46 compute_masks=compute_masks,
47 do_3D=True,
48 omni=omni,
49 flow_factor=flow_factor)
2023-08-08 01:01:25,558 [INFO] ** TORCH GPU version installed and working. **
2023-08-08 01:01:25,558 [INFO] >>plant_cp<< model set to be used
2023-08-08 01:01:25,559 [INFO] ** TORCH GPU version installed and working. **
2023-08-08 01:01:25,560 [INFO] >>>> using GPU
2023-08-08 01:01:25,643 [INFO] using dataparallel
2023-08-08 01:01:25,682 [INFO] multi-stack tiff read in as having 162 planes 1 channels
2023-08-08 01:01:26,362 [INFO] running YX: 162 planes of size (207, 443)
2023-08-08 01:01:26,390 [INFO] 0%| | 0/15 [00:00<?, ?it/s]
2023-08-08 01:01:26,550 [INFO] 7%|6 | 1/15 [00:00<00:02, 6.28it/s]
2023-08-08 01:01:26,700 [INFO] 13%|#3 | 2/15 [00:00<00:01, 6.50it/s]
2023-08-08 01:01:26,850 [INFO] 20%|## | 3/15 [00:00<00:01, 6.58it/s]
2023-08-08 01:01:27,000 [INFO] 27%|##6 | 4/15 [00:00<00:01, 6.62it/s]
2023-08-08 01:01:27,149 [INFO] 33%|###3 | 5/15 [00:00<00:01, 6.64it/s]
2023-08-08 01:01:27,298 [INFO] 40%|#### | 6/15 [00:00<00:01, 6.66it/s]
2023-08-08 01:01:27,448 [INFO] 47%|####6 | 7/15 [00:01<00:01, 6.67it/s]
2023-08-08 01:01:27,597 [INFO] 53%|#####3 | 8/15 [00:01<00:01, 6.68it/s]
2023-08-08 01:01:27,747 [INFO] 60%|###### | 9/15 [00:01<00:00, 6.68it/s]
2023-08-08 01:01:27,896 [INFO] 67%|######6 | 10/15 [00:01<00:00, 6.69it/s]
2023-08-08 01:01:28,046 [INFO] 73%|#######3 | 11/15 [00:01<00:00, 6.69it/s]
2023-08-08 01:01:28,197 [INFO] 80%|######## | 12/15 [00:01<00:00, 6.66it/s]
2023-08-08 01:01:28,347 [INFO] 87%|########6 | 13/15 [00:01<00:00, 6.66it/s]
2023-08-08 01:01:28,497 [INFO] 93%|#########3| 14/15 [00:02<00:00, 6.67it/s]
2023-08-08 01:01:28,641 [INFO] 100%|##########| 15/15 [00:02<00:00, 6.74it/s]
2023-08-08 01:01:28,642 [INFO] 100%|##########| 15/15 [00:02<00:00, 6.66it/s]
2023-08-08 01:01:28,868 [INFO] running ZY: 207 planes of size (162, 443)
2023-08-08 01:01:28,900 [INFO] 0%| | 0/19 [00:00<?, ?it/s]
2023-08-08 01:01:29,030 [INFO] 5%|5 | 1/19 [00:00<00:02, 7.77it/s]
2023-08-08 01:01:29,159 [INFO] 11%|# | 2/19 [00:00<00:02, 7.76it/s]
2023-08-08 01:01:29,288 [INFO] 16%|#5 | 3/19 [00:00<00:02, 7.75it/s]
2023-08-08 01:01:29,417 [INFO] 21%|##1 | 4/19 [00:00<00:01, 7.75it/s]
2023-08-08 01:01:29,546 [INFO] 26%|##6 | 5/19 [00:00<00:01, 7.75it/s]
2023-08-08 01:01:29,675 [INFO] 32%|###1 | 6/19 [00:00<00:01, 7.75it/s]
2023-08-08 01:01:29,806 [INFO] 37%|###6 | 7/19 [00:00<00:01, 7.71it/s]
2023-08-08 01:01:29,935 [INFO] 42%|####2 | 8/19 [00:01<00:01, 7.71it/s]
2023-08-08 01:01:30,065 [INFO] 47%|####7 | 9/19 [00:01<00:01, 7.72it/s]
2023-08-08 01:01:30,194 [INFO] 53%|#####2 | 10/19 [00:01<00:01, 7.73it/s]
2023-08-08 01:01:30,323 [INFO] 58%|#####7 | 11/19 [00:01<00:01, 7.74it/s]
2023-08-08 01:01:30,452 [INFO] 63%|######3 | 12/19 [00:01<00:00, 7.74it/s]
2023-08-08 01:01:30,582 [INFO] 68%|######8 | 13/19 [00:01<00:00, 7.73it/s]
2023-08-08 01:01:30,712 [INFO] 74%|#######3 | 14/19 [00:01<00:00, 7.72it/s]
2023-08-08 01:01:30,841 [INFO] 79%|#######8 | 15/19 [00:01<00:00, 7.73it/s]
2023-08-08 01:01:30,970 [INFO] 84%|########4 | 16/19 [00:02<00:00, 7.73it/s]
2023-08-08 01:01:31,099 [INFO] 89%|########9 | 17/19 [00:02<00:00, 7.74it/s]
2023-08-08 01:01:31,228 [INFO] 95%|#########4| 18/19 [00:02<00:00, 7.74it/s]
2023-08-08 01:01:31,354 [INFO] 100%|##########| 19/19 [00:02<00:00, 7.79it/s]
2023-08-08 01:01:31,355 [INFO] 100%|##########| 19/19 [00:02<00:00, 7.74it/s]
2023-08-08 01:01:31,694 [INFO] running ZX: 443 planes of size (162, 207)
2023-08-08 01:01:31,732 [INFO] 0%| | 0/14 [00:00<?, ?it/s]
2023-08-08 01:01:31,853 [INFO] 7%|7 | 1/14 [00:00<00:01, 8.28it/s]
2023-08-08 01:01:31,974 [INFO] 14%|#4 | 2/14 [00:00<00:01, 8.25it/s]
2023-08-08 01:01:32,096 [INFO] 21%|##1 | 3/14 [00:00<00:01, 8.24it/s]
2023-08-08 01:01:32,219 [INFO] 29%|##8 | 4/14 [00:00<00:01, 8.21it/s]
2023-08-08 01:01:32,341 [INFO] 36%|###5 | 5/14 [00:00<00:01, 8.21it/s]
2023-08-08 01:01:32,462 [INFO] 43%|####2 | 6/14 [00:00<00:00, 8.22it/s]
2023-08-08 01:01:32,584 [INFO] 50%|##### | 7/14 [00:00<00:00, 8.22it/s]
2023-08-08 01:01:32,705 [INFO] 57%|#####7 | 8/14 [00:00<00:00, 8.22it/s]
2023-08-08 01:01:32,827 [INFO] 64%|######4 | 9/14 [00:01<00:00, 8.22it/s]
2023-08-08 01:01:32,948 [INFO] 71%|#######1 | 10/14 [00:01<00:00, 8.23it/s]
2023-08-08 01:01:33,070 [INFO] 79%|#######8 | 11/14 [00:01<00:00, 8.22it/s]
2023-08-08 01:01:33,191 [INFO] 86%|########5 | 12/14 [00:01<00:00, 8.22it/s]
2023-08-08 01:01:33,313 [INFO] 93%|#########2| 13/14 [00:01<00:00, 8.22it/s]
2023-08-08 01:01:33,432 [INFO] 100%|##########| 14/14 [00:01<00:00, 8.28it/s]
2023-08-08 01:01:33,432 [INFO] 100%|##########| 14/14 [00:01<00:00, 8.23it/s]
2023-08-08 01:01:34,940 [INFO] network run in 9.22s
dP_ times 10 for >2d, still experimenting
2023-08-08 01:01:38,310 [INFO] masks created in 3.37s
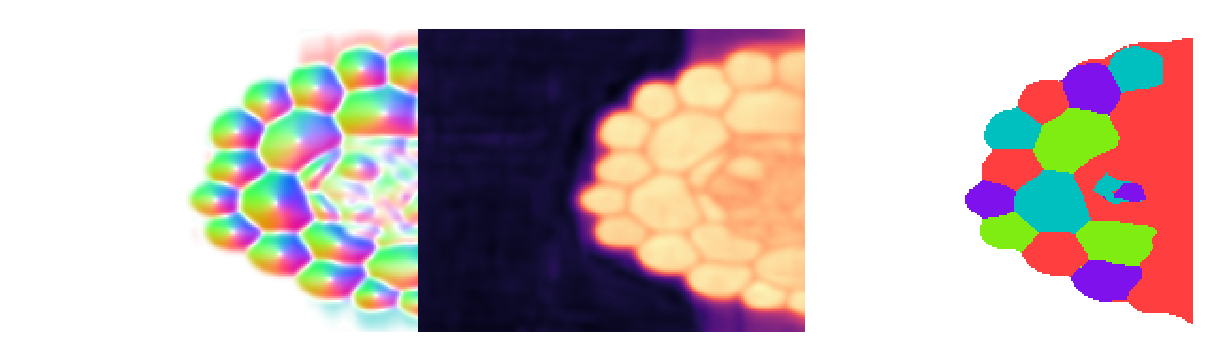
Compare masks to ground truth#
1from fastremap import unique
2mgt = io.imread(files[0][:-4]+'_masks.tif')
3print('Cellpose do_3D + omni=True: {} masks. \nOmnipose 3D: {} masks. \nGround truth: {} masks'.format(len(unique(masks_cp[0])),
4 len(unique(masks_om[0])),
5 len(unique(mgt))))
Cellpose do_3D + omni=True: 55 masks.
Omnipose 3D: 204 masks.
Ground truth: 67 masks
For what it's worth, pure Cellpose gives ~550 masks in this volume, pure Omnipose gives ~200, and Cellpose model + Omnipose mask reconstruction gives ~50. I'm sorry to say that the ground truth for this dataset is quite bad, containing some undersegmented cells, but more importantly, an entire "ignore" region where there are many, many cells that are unlabeled. So the count of 67 cells in the ground truth refers only to the long cells on the outside of the root. Thus, 55 cells is a severe under-segmentation of the volume. Let's see why.
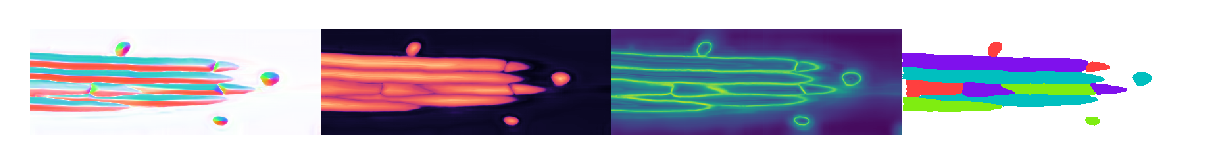
Plot results#
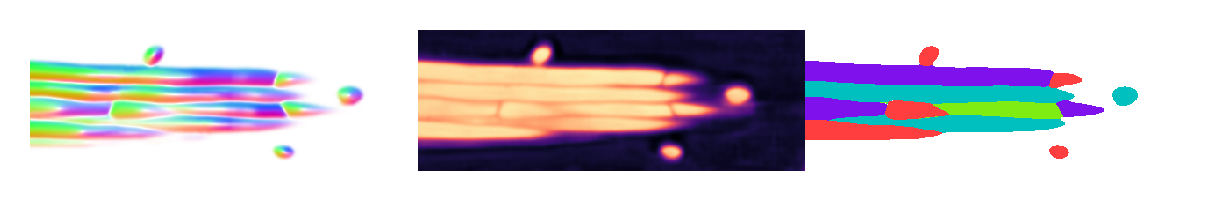
1from cellpose_omni import plot
2from omnipose.plot import apply_ncolor
3
4mu = flows_cp[0][1]
5T = flows_cp[0][2]
6bd = flows_cp[0][4]
7# mu.shape,T.shape,bd.shape
8
9d = mu.shape[0]
10
11from omnipose.utils import rescale
12c = np.array([1]*2+[0]*(d-2))
13# c = np.arange(d)
14def cyclic_perm(a):
15 n = len(a)
16 b = [[a[i - j] for i in range(n)] for j in range(n)]
17 return b
18slices = []
19idx = np.arange(d)
20cmap = mpl.colormaps['magma']
21cmap2 = mpl.colormaps['viridis']
22
23for inds in cyclic_perm(c):
24 slc = tuple([slice(-1) if i else mu.shape[k+1]//2 for i,k in zip(inds,idx)])
25 flow = plot.dx_to_circ(mu[np.where(inds)+slc],transparency=1)/255
26 dist = cmap(rescale(T)[slc])
27 msks = apply_ncolor(masks_cp[0][slc])
28
29 fig = plt.figure(figsize=[5]*2,frameon=False)
30 plt.imshow(np.hstack((flow,dist,msks)),interpolation='none')
31
32 plt.axis('off')
33 plt.show()
1%%capture
2import ncolor
3mask = masks_cp[0]
4mask_nc = ncolor.label(mask,max_depth=20)
5
6import napari
7viewer = napari.view_labels(mask_nc);
8viewer.dims.ndisplay = 3
9viewer.camera.center = [s//2 for s in mask.shape]
10viewer.camera.zoom=1
11viewer.camera.angles=(10.90517458968619, -20.777067798396835, 58.04311170773853)
12viewer.camera.perspective=0.0
13viewer.camera.interactive=True
14
15img = viewer.screenshot(size=(1000,1000),scale=1,canvas_only=True,flash=False)
1plt.figure(figsize=(3,3),frameon=False)
2plt.imshow(img)
3plt.axis('off')
4plt.show()

It appears that omni=True does allow 2D Cellpose models to work in 3D, but the prediction quality - worsened by artifacts introduced by the compoisiting into 3D - is a limiting factor.