Affinity segmentation#
This is the term that I think best describes encoding an image segmentation it its most general, information-dense form: an affinity graph. To explain what this is, we will first consider two cells in contact.
The hierarchy of segmentation encoding#
Show code cell source
1# Load image and masks
2import string
3import matplotlib as mpl
4import matplotlib.pyplot as plt
5mpl.rcParams['figure.dpi'] = 600
6# mpl.rcParams['facecolor'] = [0]*4
7
8plt.rc('figure', facecolor=[0]*4)
9
10plt.style.use('dark_background')
11mpl.use('Agg')
12
13%matplotlib inline
14
15from pathlib import Path
16import os
17from cellpose_omni import io, plot
18import fastremap
19
20import omnipose
21omnidir = Path(omnipose.__file__).parent.parent
22basedir = os.path.join(omnidir,'docs','_static')
23# name = 'ecoli_phase'
24name = 'ecoli'
25ext = '.tif'
26image = io.imread(os.path.join(basedir,name+ext))
27masks = io.imread(os.path.join(basedir,name+'_labels'+ext))
28slc = omnipose.utils.crop_bbox(masks>0,pad=0)[0]
29masks = fastremap.renumber(masks[slc])[0]
30image = image[slc]
31
32# Plot a few things
33
34import matplotlib.pyplot as plt
35from omnipose.plot import apply_ncolor, plot_edges, imshow
36from omnipose import utils
37import numpy as np
38# import matplotlib_inline
39# matplotlib_inline.backend_inline.set_matplotlib_formats('svg')
40
41from mpl_toolkits.axes_grid1.inset_locator import zoomed_inset_axes
42from mpl_toolkits.axes_grid1.inset_locator import inset_axes
43from mpl_toolkits.axes_grid1.inset_locator import mark_inset
44from matplotlib.patches import Patch
45
46
47
48images = [image, masks>0, apply_ncolor(masks), masks>0]
49labels = ['Image\n(phase contrast)', 'Semantic\nsegmentation',
50 'Instance\nsegmentation', 'Affinity\nsegmentation']
51
52# Set up the figure and subplots
53N = len(images)
54
55f = 1
56Y,X = masks.shape[-2:]
57M = 1
58
59h,w = masks.shape[-2:]
60
61sf = w
62p = 0.0035*w # needs to be defined as fraction of width for aspect ratio to work?
63h /= sf
64w /= sf
65
66# Calculate positions of subplots
67left = np.array([i*(w+p) for i in range(N)])*1.
68bottom = np.array([0]*N)*1.
69width = np.array([w]*N)*1.
70height = np.array([h]*N)*1.
71
72max_w = left[-1]+width[-1]
73max_h = bottom[-1]+height[-1]
74
75sw = max_w
76sh = max_h
77
78sf = max(sw,sh)
79left /= sw
80bottom /= sh
81width /= sw
82height /= sh
83
84# Create figure
85s = 6
86fig = plt.figure(figsize=(s,s*sh/sw), frameon=False, dpi=300)
87# fig.patch.set_facecolor([0]*4)
88
89# Add subplots
90axes = []
91for i in range(N):
92 ax = fig.add_axes([left[i], bottom[i], width[i], height[i]])
93 axes.append(ax)
94
95# Iterate over each subplot and set the image, label, and formatting
96c = [0.5]*3
97fontsize = 11
98
99# bounds = [40,20,10,10]
100bounds = [11,22,8,8]
101h,w = masks.shape[-2:]
102extent = np.array([0,w,0,h])#-0.5
103
104
105sy,sx,wy,wx = bounds
106zoomslc = tuple([slice(sy,sy+wy),slice(sx,sx+wx)])
107
108
109cmap='inferno'
110
111zoom = 3
112# zoom, f = 5, 0.75
113color = [.75]*3
114edgecol = [1/3]*3
115edgecol = [.75]*3+[.5]
116axcol = [0.5]*3
117# edgecol = [.5,.75,0]+[2/3]
118
119lw= .2
120labelpad = 3
121fontsize2 = 8
122
123do_labels = 0
124
125for i, ax in enumerate(axes):
126
127 # inset axes
128 axins = zoomed_inset_axes(ax, zoom, loc='lower left',
129 bbox_to_anchor=(-wx/w,-2*wy/h),
130 # bbox_to_anchor=(-wx/w*zoom/2,-zoom*wy/h),
131 # bbox_to_anchor=(-f*zoom*wy/h,-f*wx/w*zoom),
132
133
134 bbox_transform=ax.transAxes)
135
136 if i==N-1:
137 # ax.invert_yaxis()
138
139 dim = masks.ndim
140 shape = masks.shape
141 steps, inds, idx, fact, sign = utils.kernel_setup(dim)
142 coords = np.nonzero(masks)
143 affinity_graph = omnipose.core.masks_to_affinity(masks, coords, steps,
144 inds, idx, fact, sign, dim)
145 neighbors = utils.get_neighbors(coords,steps,dim,shape)
146 summed_affinity, affinity_cmap = plot_edges(shape,affinity_graph,neighbors,coords,
147 figsize=1,fig=fig,ax=ax,extent=extent,
148 edgecol=edgecol,cmap=cmap,linewidth=lw
149 )
150
151
152 axins.invert_yaxis()
153 ax.invert_yaxis()
154
155 summed_affinity, affinity_cmap = plot_edges(shape,affinity_graph,neighbors,coords,
156 figsize=1,fig=fig,ax=axins,
157 extent=extent,
158 edgecol=edgecol,linewidth=lw*zoom,cmap=cmap,
159 bounds=bounds
160 )
161
162 axins.set_xlim(zoomslc[1].start, zoomslc[1].stop)
163 axins.set_ylim(h-zoomslc[0].start, h-zoomslc[0].stop)
164
165
166 loc1,loc2 = 4,2
167 patch, pp1, pp2 = mark_inset(ax, axins, loc1=loc1, loc2=loc2, fc="none", ec=color+[1],zorder=2)
168 pp1.loc1 = 4
169 pp1.loc2 = 1
170 pp2.loc1 = 2
171 pp2.loc2 = 3
172
173
174 N = affinity_cmap.N
175 colors = affinity_cmap.colors
176
177 cax = inset_axes(ax, width="50%", height="100%", loc='lower right',
178 bbox_to_anchor=(-.05, -0.7, 1, 1), bbox_transform=ax.transAxes,
179 borderpad=0)
180
181 # Display the color swatches as an image
182 n = np.arange(3,9)
183 Nc = len(n)
184 cax.imshow(affinity_cmap(n.reshape(1,Nc)))#,vmin=n[0]-1,vmax=n[-1]+1)
185
186 # Set the y ticks and tick labels
187 cax.set_xticks(np.arange(Nc))
188 nums = [str(i) for i in n]
189 cax.set_xticklabels(nums,c=c,fontsize=fontsize2)
190 cax.tick_params(axis='both', which='both', length=0, pad=labelpad)
191 cax.set_yticks([])
192
193 wa = .07
194 ha = .05
195 cax.set_aspect(ha/wa)
196 cax.set_title('Connections',c=c,fontsize=fontsize2,pad=labelpad)
197 for spine in cax.spines.values():
198 spine.set_color(None)
199 else:
200
201 ax.imshow(images[i],cmap='gray',extent=extent)
202 # axins.imshow(images[i][zoomslc],extent=extent,origin='upper')
203 axins.imshow(images[i],extent=extent,cmap='gray')
204 # axins.imshow(images[i])#,extent=extent)
205
206 if i>0:
207 imp = masks[::-1][zoomslc]
208 if i==1:
209 imp = imp>0
210 for (j,k),label in np.ndenumerate(imp):
211 axins.text(k+sx+0.5, j+sy+0.45, int(label), ha='center', va='center', color=[(label==0)*0.5]*3,fontsize=4)
212
213
214 axins.set_xlim(zoomslc[1].start, zoomslc[1].stop)
215 axins.set_ylim(zoomslc[0].start, zoomslc[0].stop)
216
217 mark_inset(ax, axins, loc1=2, loc2=4, fc="none", ec=color+[1],zorder=2)
218
219
220 if do_labels:
221 ax.set_title(labels[i],c=c,fontsize=fontsize,fontweight="bold",pad=5)
222 else:
223 ax.annotate(string.ascii_lowercase[i], xy=(-0.1, 1), xycoords='axes fraction',
224 xytext=(0, 0), textcoords='offset points', va='top', c=axcol,
225 fontsize=fontsize)
226
227 ax.axis('off')
228 axins.set_xticks([])
229 axins.set_yticks([])
230 axins.set_facecolor([0]*4)
231
232 for spine in axins.spines.values():
233 spine.set_color(color)
234
235
236# plt.subplots_adjust(wspace=0.15)
237fig.patch.set_facecolor([0]*4)
238
239# Display the plot
240plt.show()
Semantic segmentation sorts pixels into semantic classes. This is often just two classes, or binary classification: foreground and background. In this case, foreground is cell and background is media. As you can see, semantic segmentation does not discern between adjacent instances of foreground objects. We store a semantic segmentation as a binary image file with the same dimensions as the image itself, with foreground pixels labeled True (1) and background False (0).
Instance segmentation assigns a unique integer to the pixels each instance of an object - in this case, each cell. This is also conveniently stored as an image file, typically uint8 (unsigned 8-bit integer) for up to \(2^{8}-1=255\) labels or uint16 (unsigned 16-bit integer) for up to \(2^{16}-1=65535\) labels. Signed and/or unsigned 32- or 64-bit formats may also be used, but your OS may not be able to preview these files in its native file manager.
Note
Some instance labels use -1 as an "ignore" label. This can be in conflict with several tasks from indexing to label formatting, which assume unsigned integers, so care must be taken when working with signed formats (int) versus unsigned (uint).
Bad labels I: Semantic islands to instance labels#
Semantic segmentation can be converted into instance segmentation, and this forms the basis of many instance segmentation pipelines. The general steps are:
Pre-process image: traditional filtering/blurring/feature extraction or DNN transformation
Threshold processed image: adaptive techniques are usually used on the pre-processed image to ensure that the majority of objects pixels are identified despite variations within an image and among images in a dataset. Importantly, object boundaries must not be identified as foreground. This allows each object to be associated with a unique island of foreground pixels.
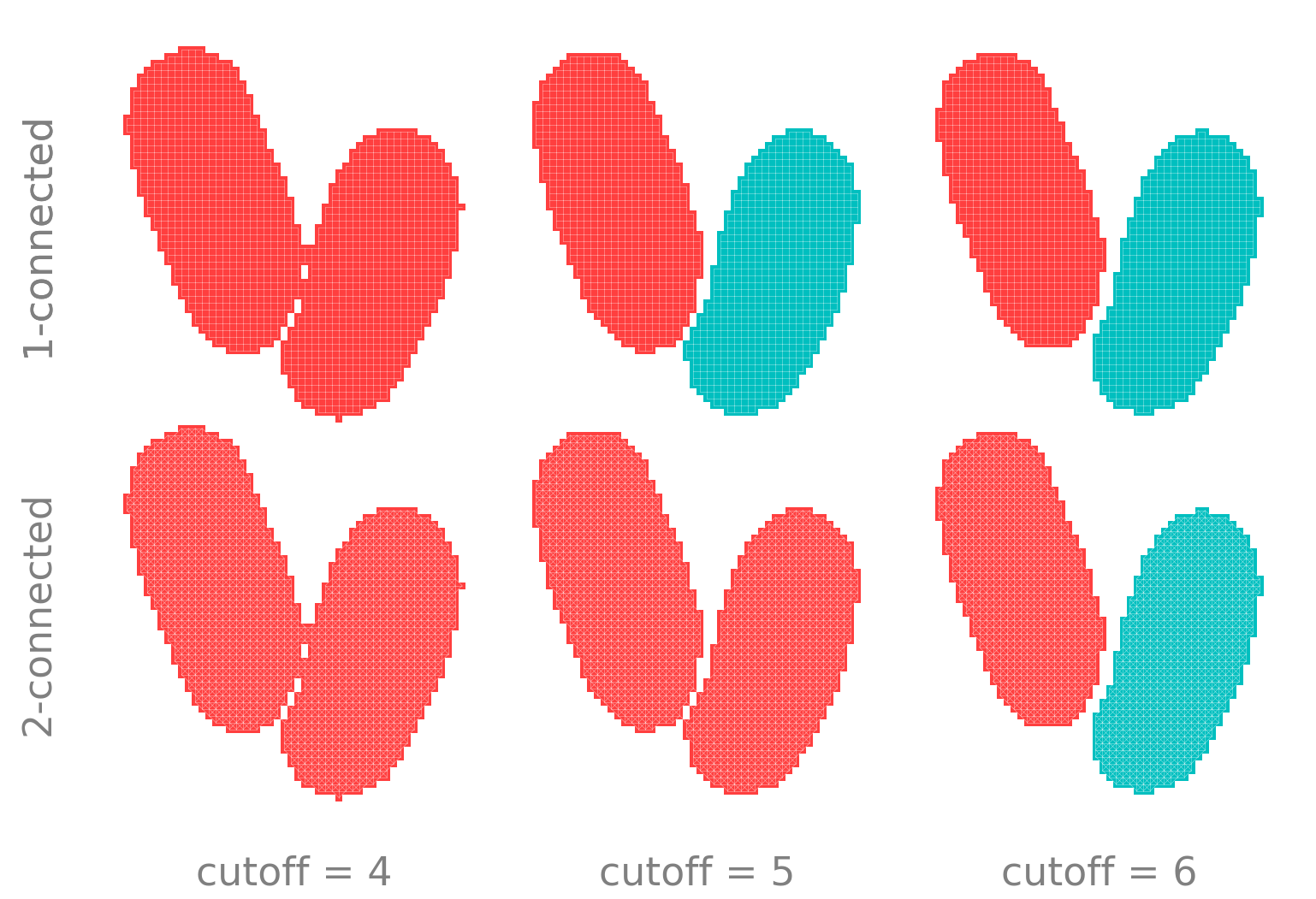
These unique blobs are identified using connected components labeling. This is the process of building an affinity graph, where pixels are nodes and edges are formed between any adjacent foreground pixels. Adjacency can be defined most narrowly by sharing edges (1-connected in Python, 4-connected in MATLAB) or more broadly by sharing either edges or vertices (2-connected in Python, 8-connected in MATLAB). The graph is then traversed to find all connected components of the graph.
These points are illustrated below. By simulating the amount of foreground pixels detected by filtering+thresholding, we see that is is impossible to distinguish between the two cells until much of the boundary is lost, particularly when using 2-connectivity.
Show code cell source
1import skimage.measure
2
3connectivity = [1,2]
4cutoffs = [4,5,6]
5row_labels = ['{}-connected'.format(i) for i in connectivity]
6col_labels = ['cutoff = {}'.format(i) for i in cutoffs]
7# Define the grid dimensions
8num_rows = len(row_labels)
9num_cols = len(col_labels)
10
11# Create the grid of subplots
12
13# f = .75
14# dpi = mpl.rcParams['figure.dpi']
15# Y,X = masks.shape[-2:]
16# szX = max(X//dpi,2)*f
17# szY = max(Y//dpi,2)*f
18# fig, axes = plt.subplots(num_rows, num_cols,figsize=(szX*num_cols,szY*num_rows))
19
20
21
22
23h,w = masks.shape[-2:]
24
25sf = w
26p = 0.05
27h /= sf
28w /= sf
29
30# Calculate positions of subplots
31N = num_cols
32M = num_rows
33left = np.array([5*p+i*(w+p) for i in range(N)]*M).flatten().astype(float)
34# bottom = np.array([1.5*p]*N + [h+p]*N).flatten().astype(float)
35bottom = np.array([h+p]*N+[3*p]*N).flatten().astype(float)
36width = np.array([[w]*N]*M).flatten().astype(float)
37height = np.array([[h]*N]*M).flatten().astype(float)
38
39max_w = left[-1]+width[-1]
40max_h = bottom[-1]+height[-1]
41
42sw = max_w
43sh = max_h
44
45sf = max(sw,sh)
46left /= sw
47bottom /= sh
48width /= sw
49height /= sh
50
51# Create figure
52s = 5
53fig = plt.figure(figsize=(s,s*sh/sw), frameon=False, dpi=300,facecolor=[0]*4)
54# fig.patch.set_facecolor([0]*4)
55
56# Add subplots
57axes = []
58for i in range(N*M):
59 ax = fig.add_axes([left[i], bottom[i], width[i], height[i]])
60 ax.set_facecolor([0]*4)
61
62 axes.append(ax)
63
64
65fig.patch.set_facecolor([0]*4)
66color = [0.5]*3
67# for row,conn in enumerate(connectivity):
68# for col,cutoff in enumerate(cutoffs):
69
70for i,ax in enumerate(axes):
71 row,col= np.unravel_index(i,(M,N))
72
73 cutoff = cutoffs[col]
74 conn = connectivity[row]
75
76
77 bin0 = summed_affinity>cutoff
78 msk0 = skimage.measure.label(bin0,connectivity=conn)
79 pic = apply_ncolor(msk0)
80
81
82 dim = masks.ndim
83 shape = masks.shape
84 steps, inds, idx, fact, sign = utils.kernel_setup(dim)
85 coords = np.nonzero(msk0)
86 affinity_graph = omnipose.core.masks_to_affinity(msk0, coords, steps,
87 inds, idx, fact, sign, dim)
88 neighbors = utils.get_neighbors(coords,steps,dim,shape)
89
90 #choose to plot cardinal connections only
91 step_inds = None if conn==2 else inds[1]
92
93 # index = np.ravel_multi_index([[row],[col]],(N,M))
94 # ax = axes[row*col]
95 ax.axis('off')
96
97 # ax.text(0,0,i)
98
99 plot_edges(shape,affinity_graph,neighbors,coords,figsize=1,extent=extent,
100 fig=fig,ax=ax,step_inds=step_inds,pic=pic,origin='lower',edgecol=[1,1,1,0.5])
101
102 ax.invert_yaxis()
103
104 if i<N:
105 ax.annotate(col_labels[i], xy=(0.5, 1), xytext=(0, ax.xaxis.labelpad),
106 xycoords=ax.xaxis.label, textcoords='offset points',
107 ha='center', va='baseline',color=color,fontsize=fontsize)
108
109 if i%M==0:
110 ax.annotate(row_labels[row], xy=(0, 0), xytext=(-ax.yaxis.labelpad + 20, 0),
111 xycoords=ax.yaxis.label, textcoords='offset points',
112 ha='right', va='center', rotation=90,color=color,fontsize=fontsize)
113
114plt.show()
Although such sloppy segmentation is good enough for some tasks, we have a better tools now. So in general, do not use image thresholding for segmentation.
Bad labels II: Watershed lines#
While we are on the topic, missing boundary pixels also frequently arise when applying the watershed transform. As usually implemented, this ubiquitous operation returns a semantic classification of an image into watershed lines and catchment basins. As you can tell by the above example, this means that distinct basins must be separated by a 1- or 2-connected watershed line, and therefore boundary pixels are always left unclassified.
There are implementations that allow users to return instance labels without the gaps let by watershed lines (e.g., skimage.segmentation.watershed), but I have yet to see a paper published using this method. Despite this fix, watershed also tends to over-segment images (even when transformed by traditional filters or DNNs). So in general, do not use watershed for instance segmentation.
Bad labels III: Self-contact boundaries#
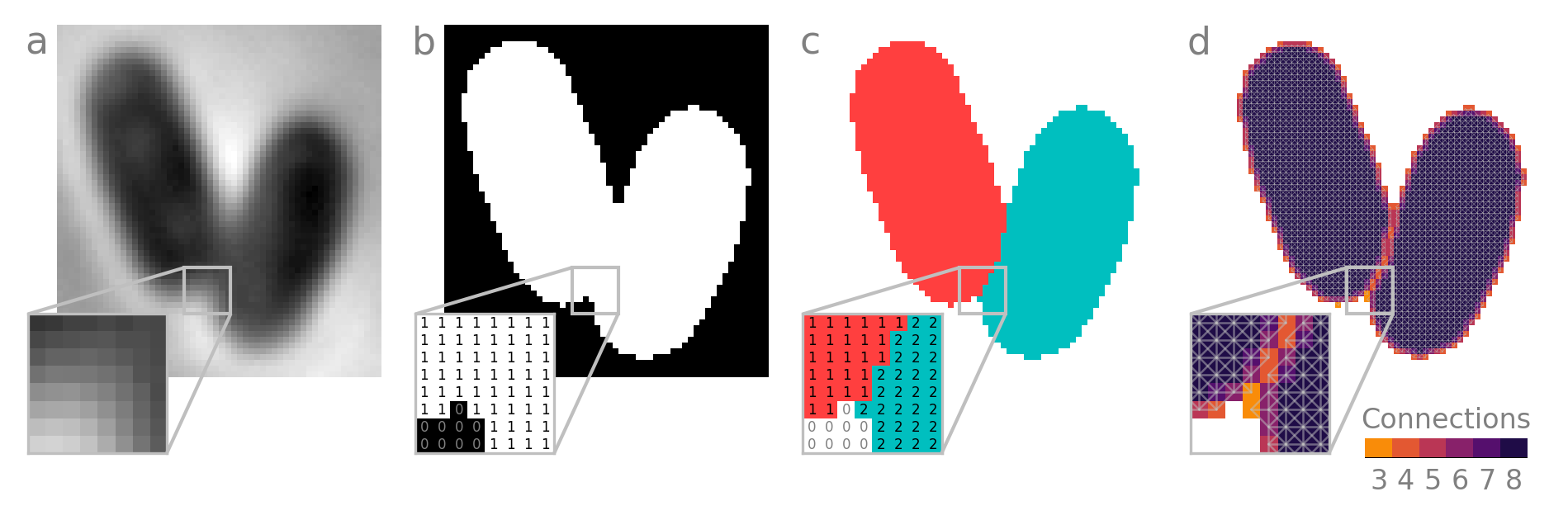
Instance labels are good enough to fully describe a lot of objects. More precisely, there is a bijective map between the affinity graph and the instance label matrix whenever all edge pixels are in contact with a non-self pixel. This assumption fails in many interesting (and biologically relevant) scenarios, including bacterial microscopy. Consider the following image containing one extremely filamentous cell and corresponding cell mask:
Show code cell source
1# read in files; this is an entire movie, but we will just be looking at the last frame
2import tifffile
3
4nm = 'long_10_2'
5masks = tifffile.imread(os.path.join(basedir,nm+'_op_masks.tif'))
6phase = tifffile.imread(os.path.join(basedir,nm+'_phase.tif'))
7fluor = tifffile.imread(os.path.join(basedir,nm+'_fluor.tif'))
8afnty = utils.load_nested_list(os.path.join(basedir,nm+'_affinity.npz'))
9
10# make figure
11import omnipose, cellpose_omni
12im = phase[-1]
13msk = masks[-1]
14
15f = 1
16c = [0.5]*3
17fontsize=11
18
19titles = [r'$\bf{image}$'+'\n(phase contrast)', r'$\bf{label}$'+'\n(single cell mask)', r'$\bf{boundary}$'+'\n(from cell mask)']
20ol = cellpose_omni.utils.masks_to_outlines(msk,omni=True)
21# outlines = np.stack([ol]*4,axis=-1)*0.5
22images = [im,
23 omnipose.plot.apply_ncolor(msk),
24 omnipose.plot.apply_ncolor(ol,offset=.5)]
25
26
27
28# Set up the figure and subplots
29N = len(images)
30h,w = im.shape
31
32sf = h
33p = 0.5 # needs to be defined as fraction of width for aspect ratio to work?
34
35
36h,w = im.shape
37extent = np.array([0,w,0,h])#-0.5
38sy,sx,wy,wx = [h//2.5,w//3.6,40,40]
39zoomslc = tuple([slice(sy,sy+wy),slice(sx,sx+wx)])
40zoom = 5
41bbox_to_anchor = (-(wx/w)*zoom/(1.25),-zoom/1.5*wy/h) # inset axis
42
43
44asp = h/w
45
46h /= sf
47w /= sf
48
49oy,ox = np.abs(bbox_to_anchor)/2
50# oy,ox = 0.,0.
51# Calculate positions of subplots
52left = np.array([ox+i*(w+p) for i in range(N)])*1.
53bottom = np.array([oy]*N)*1.
54width = np.array([w]*N)*1.
55height = np.array([h]*N)*1.
56
57max_w = left[-1]+width[-1]+ox
58max_h = bottom[-1]+height[-1]+oy
59
60sw = max_w
61sh = max_h
62
63sf = max(sw,sh)
64left /= sw
65bottom /= sh
66width /= sw
67height /= sh
68
69# Create figure
70s = 6.5
71fig = plt.figure(figsize=(s,s*sh/sw), frameon=False, dpi=600)
72# fig.patch.set_facecolor([0]*4)
73
74# Add subplots
75axes = []
76for i in range(N):
77 ax = fig.add_axes([left[i], bottom[i], width[i], height[i]])
78 axes.append(ax)
79
80
81
82lwa = 2/N # linewidth for axes
83lw = lwa/20 # linewidth for affinity graph
84labelpad = 2
85
86for i, ax in enumerate(axes):
87
88 ax.imshow(images[i],cmap='gray',extent=extent)
89 ax.axis('off')
90
91
92
93 # inset axes....
94 # axins = ax.inset_axes([0.5, 0.5, 0.47, 0.47])
95 # axins = inset_axes(ax, 1,1 , loc=2, bbox_to_anchor=(.08, 0.35))
96 axins = zoomed_inset_axes(ax, zoom, loc='lower left',
97 # bbox_to_anchor=(1.1, 1.1),
98 # bbox_to_anchor=(-.15,-.15),
99 bbox_to_anchor=bbox_to_anchor,
100 bbox_transform=ax.transAxes)
101
102
103
104 # axins.imshow(images[i][zoomslc],extent=extent,origin='upper')
105 axins.imshow(images[i],extent=extent,cmap='gray')
106 # axins.imshow(images[i])#,extent=extent)
107 axins.set_xlim(zoomslc[1].start, zoomslc[1].stop)
108 axins.set_ylim(zoomslc[0].start, zoomslc[0].stop)
109
110 mark_inset(ax, axins, loc1=2, loc2=4, fc="none", ec=color+[1], zorder=2, lw=lwa)
111 # axins.axis('off')
112 axins.set_xticks([])
113 axins.set_yticks([])
114 axins.set_facecolor([0]*4)
115
116
117 for spine in axins.spines.values():
118 spine.set_color(color)
119 spine.set_linewidth(lwa)
120
121
122 if do_labels:
123 # ax.set_title(labels[i],c=c,fontsize=fontsize,fontweight="bold",pad=5)
124 ax.set_title(titles[i],c=c,fontsize=fontsize,pad=5)
125
126 else:
127 ax.annotate(string.ascii_lowercase[i], xy=(-0.25, 1), xycoords='axes fraction',
128 xytext=(0, 0), textcoords='offset points', va='top', c=axcol,
129 fontsize=fontsize)
130
131
132
133# plt.subplots_adjust(wspace=0,hspace=0)
134
135# Display the plot
136plt.show()
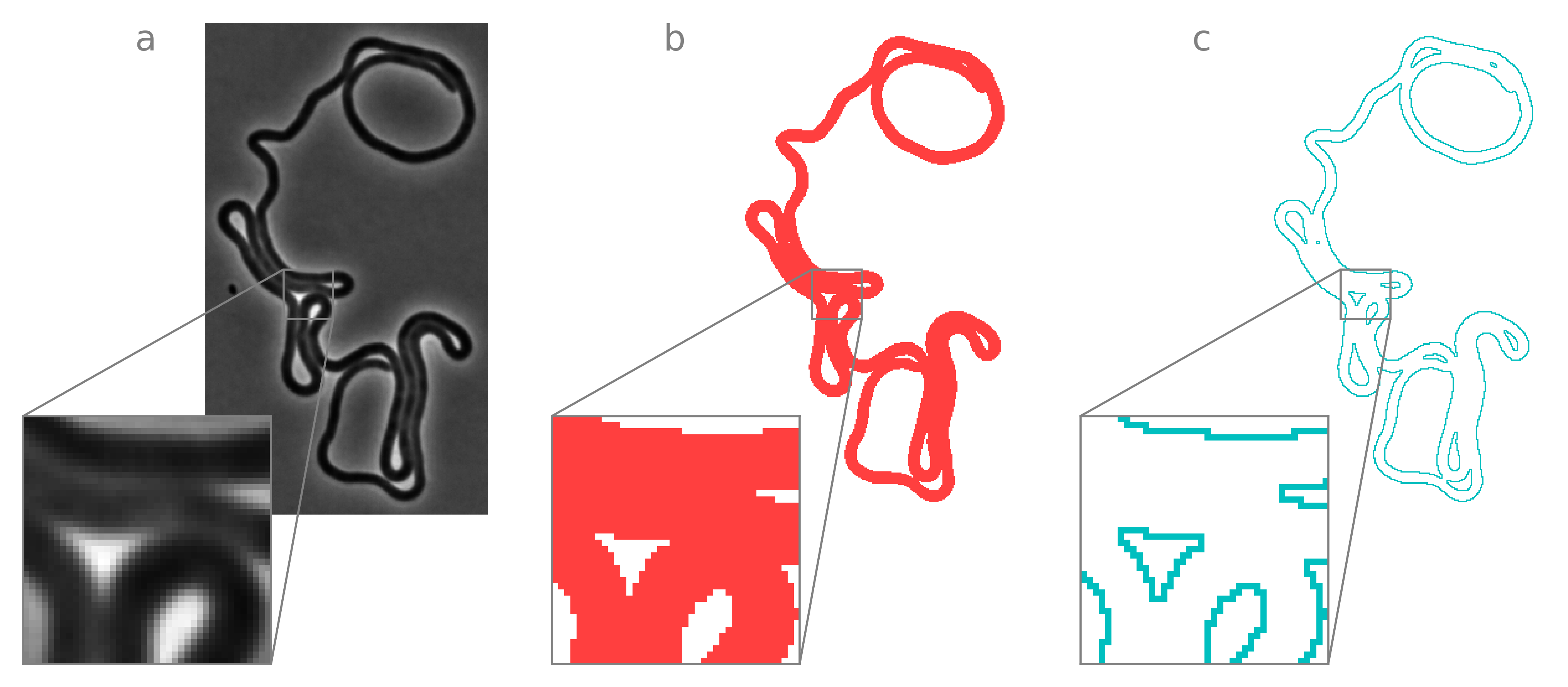
Because the label for this cell is the same integer on either side of a self-contact interface, we cannot localize the boundary of the cell at these interfaces. However, affinity segmentation encodes not only the information necessary to recontruct cell boundaries but also to traverse cell boundarues as a parametric contour.
Show code cell source
1import omnipose, cellpose_omni
2from scipy import signal
3t = -1 # last frame
4im = phase[t]
5msk = masks[t]
6
7f = 1
8c = [0.5]*3
9fontsize = 11
10
11titles = [#r'$\bf{image}$'+'\n(phase contrast)',
12 r'$\bf{connectivity}$'+'\n(affinity graph)',
13 r'$\bf{boundary}$'+'\n(from affinity graph)',
14 r'$\bf{contour}$'+'\n(traced with affinity)']
15
16# extract the addinity graph and coordinate array
17aa = afnty[t]
18shape = msk.shape
19dim = msk.ndim
20neighbors = aa[:dim]
21affinity_graph = aa[dim]#.astype(bool) #VERY important to cast to bool, now done internally
22idx = affinity_graph.shape[0]//2
23coords = tuple(neighbors[:,idx])
24
25# make the boundary
26ol = omnipose.core.affinity_to_boundary(msk,affinity_graph,coords)
27
28# make the contour
29contour_map, contour_list, unique_L = omnipose.core.get_contour(msk,affinity_graph,coords,cardinal_only=1)
30
31cmap='inferno'
32color = [0.5]*3
33
34
35
36# contour_colored = np.stack([(contour_map>1).astype(np.float32)]*4,axis=-1)
37contour_colored = np.zeros(contour_map.shape+(4,))
38
39for contour in contour_list:
40 # coords_t = np.unravel_index(contour,contour_map.shape)
41 coords_t = np.stack([c[contour] for c in coords])
42 cyclic_diff = np.diff(np.append(coords_t, coords_t[:, 0:1], axis=1), axis=1)
43
44 a = cyclic_diff
45 window_size = 11
46 window = np.ones(window_size) / window_size
47 cyclic_diff = signal.convolve2d(np.concatenate((a[:, -window_size+1:], a, a[:, :window_size-1]), axis=1), np.expand_dims(window, axis=0), mode='same')[:, window_size-1:-window_size+1]
48
49 angles = np.arctan2(cyclic_diff[1], cyclic_diff[0])+np.pi
50
51
52 a = 2
53 r = ((np.cos(angles)+1)/a)
54 g = ((np.cos(angles+2*np.pi/3)+1)/a)
55 b =((np.cos(angles+4*np.pi/3)+1)/a)
56
57 rgb = np.stack((r,g,b,np.ones_like(angles)),axis=-1)
58
59 # v = np.array(range(len(contour)))/len(contour)
60 # contour_colored[tuple(coords_t)] = ctr_cmap(v)
61 contour_colored[tuple(coords_t)] = rgb
62
63
64images = [#im,
65 None,
66 omnipose.plot.apply_ncolor(ol,offset=.5),
67 contour_colored]
68
69# N = len(images)
70# A = N//2
71# B = N-A
72
73# fig, axes = plt.subplots(2,B, figsize=(szX*A,szY*B))
74# fig.patch.set_facecolor([0]*4)
75
76# inset axis
77
78
79# Set up the figure and subplots
80N = len(images)
81h,w = im.shape
82
83
84extent = np.array([0,w,0,h])#-0.5
85sy,sx,wy,wx = [h//2.5,w//3.6,40,40]
86zoomslc = tuple([slice(sy,sy+wy),slice(sx,sx+wx)])
87zoom = 5
88bbox_to_anchor = (-(wx/w)*zoom/(1.25),-zoom/1.5*wy/h)
89asp = h/w
90
91sf = h
92p = 0.5 # needs to be defined as fraction of width for aspect ratio to work?
93h /= sf
94w /= sf
95
96oy,ox = np.abs(bbox_to_anchor)/2
97# oy,ox = 0.,0.
98# Calculate positions of subplots
99left = np.array([ox+i*(w+p) for i in range(N)])*1.
100bottom = np.array([oy]*N)*1.
101width = np.array([w]*N)*1.
102height = np.array([h]*N)*1.
103
104max_w = left[-1]+width[-1]+ox
105max_h = bottom[-1]+height[-1]+oy
106
107sw = max_w
108sh = max_h
109
110sf = max(sw,sh)
111left /= sw
112bottom /= sh
113width /= sw
114height /= sh
115
116# Create figure
117s = 6.5
118fig = plt.figure(figsize=(s,s*sh/sw), frameon=False, dpi=600)
119
120# Add subplots
121axes = []
122for i in range(N):
123 ax = fig.add_axes([left[i], bottom[i], width[i], height[i]])
124 axes.append(ax)
125
126
127
128h,w = im.shape
129
130
131lwa = 2/N # linewidth for axes
132lw = lwa/20 # linewidth for affinity graph
133labelpad = 4/3
134
135fontsize2 = 8
136
137for i, ax in enumerate(axes):
138
139 # inset axes....
140 # axins = ax.inset_axes([0.5, 0.5, 0.47, 0.47])
141 # axins = inset_axes(ax, 1,1 , loc=2, bbox_to_anchor=(.08, 0.35))
142 axins = zoomed_inset_axes(ax, zoom, loc='lower left',
143 # bbox_to_anchor=(1.1, 1.1),
144 # bbox_to_anchor=(-.15,-.15),
145 bbox_to_anchor=bbox_to_anchor,
146 bbox_transform=ax.transAxes)
147
148 if i==(N-3):
149 # ax.invert_yaxis()
150 neighbors = utils.get_neighbors(coords,steps,dim,shape)
151
152 # plot the while affinity graph
153 summed_affinity, affinity_cmap = plot_edges(shape,affinity_graph,neighbors,coords,
154 figsize=1,fig=fig,ax=ax,extent=extent,
155 edgecol=edgecol,cmap=cmap,linewidth=lw
156
157 )
158
159 # plot the inset one
160 axins.invert_yaxis()
161 ax.invert_yaxis()
162
163 summed_affinity, affinity_cmap = plot_edges(shape,affinity_graph,neighbors,coords,
164 figsize=1,fig=fig,ax=axins,
165 extent=extent,
166 edgecol=edgecol,linewidth=lw*zoom,cmap=cmap
167 )
168
169 # axins.set_xlim(zoomslc[1].start, zoomslc[1].stop)
170 axins.set_xlim(zoomslc[1].start, zoomslc[1].stop)
171
172 # axins.set_ylim(zoomslc[0].stop, zoomslc[0].start)
173 # axins.set_ylim(h-zoomslc[0].stop, h-zoomslc[0].start)
174 axins.set_ylim(h-zoomslc[0].start, h-zoomslc[0].stop)
175
176
177 loc1,loc2 = 4,2
178 patch, pp1, pp2 = mark_inset(ax, axins, loc1=loc1, loc2=loc2, fc="none",
179 ec=color+[1],zorder=2,lw=lwa)
180 pp1.loc1 = 4
181 pp1.loc2 = 1
182 pp2.loc1 = 2
183 pp2.loc2 = 3
184
185
186
187
188 # Display the color swatches as an image
189 Nc = affinity_cmap.N
190 colors = affinity_cmap.colors
191
192 wa = .07
193 ha = .05
194 # cax = fig.add_axes([.3975, -.08, wa, ha])
195 cax = inset_axes(ax, width="60%", height="100%", loc='lower right',
196 bbox_to_anchor=(0, -0.725, 1, 1), bbox_transform=ax.transAxes,
197 borderpad=0)
198
199 n = np.arange(3,9)
200 Nc = len(n)
201 cax.imshow(affinity_cmap(n.reshape(1,Nc)))#,vmin=n[0]-1,vmax=n[-1]+1)
202
203 # Set the y ticks and tick labels
204 # cax.set_yticks(np.arange(N))
205 cax.set_xticks(np.arange(Nc))
206
207 nums = [str(i) for i in n]
208 cax.set_xticklabels(nums,c=c,fontsize=fontsize2)
209 cax.tick_params(axis='both', which='both', length=0, pad=labelpad)
210 cax.set_yticks([])
211
212 wa = .07
213 ha = .05
214 cax.set_aspect(ha/wa)
215 cax.set_title('Connections',c=c,fontsize=fontsize2, pad=labelpad)
216 for spine in cax.spines.values():
217 spine.set_color(None)
218
219 # cax.xaxis.set_labelpad = -10
220 else:
221
222
223
224 ax.imshow(images[i],cmap='gray',extent=extent)
225 # axins.imshow(images[i][zoomslc],extent=extent,origin='upper')
226 axins.imshow(images[i],extent=extent,cmap='gray')
227 # axins.imshow(images[i])#,extent=extent)
228 axins.set_xlim(zoomslc[1].start, zoomslc[1].stop)
229 axins.set_ylim(zoomslc[0].start, zoomslc[0].stop)
230
231 mark_inset(ax, axins, loc1=2, loc2=4, fc="none",
232 ec=color+[1], zorder=2, lw=lwa)
233
234
235
236 if i==(N-1):
237 # ax2 = fig.add_axes([.7, -.15, .25, .25])
238 ax2 = inset_axes(ax, width="60%", height="100%", loc='lower right',
239 bbox_to_anchor=(0, -0.675, 1, 1), bbox_transform=ax.transAxes,
240 borderpad=0)
241 lw2 = 1
242
243 # Create the circle and arrows on the second subplot
244 circle = plt.Circle((0, 0), 1, fill=False, edgecolor=c,lw=lw2)
245 ax2.add_artist(circle)
246
247 # Set the number of arrows and colormap
248 n_arrows = 11
249 cmap = plt.get_cmap('hsv')
250
251 for j in range(n_arrows):
252 angle = j * 2 * np.pi / n_arrows
253 a = 2
254 r = ((np.cos(angle)+1)/a)
255 g = ((np.cos(angle+2*np.pi/3)+1)/a)
256 b =((np.cos(angle+4*np.pi/3)+1)/a)
257
258 rgb = np.stack((r,g,b,np.ones_like(angle)),axis=-1)
259
260
261 x, y = np.cos(angle), np.sin(angle)
262 # dx, dy = -y, x
263 dx, dy = y,-x
264
265 # clr = cmap(j / n_arrows)
266 ax2.quiver(x, y, dx, dy, color=rgb, angles='xy', scale_units='xy', scale=2, width=.05)
267
268 # Add text to the center of the circle
269 ax2.text(0, 0, 'Angle', ha='center', va='center',c=c,fontsize=fontsize2)
270
271 # Set the axis limits and aspect ratio
272 ax2.set_xlim(-1.5, 1.5)
273 ax2.set_ylim(-1.5, 1.5)
274 ax2.set_aspect('equal')
275
276 # Remove the axes from the second subplot
277 ax2.axis('off')
278
279
280 if do_labels:
281 # ax.set_title(labels[i],c=c,fontsize=fontsize,fontweight="bold",pad=5)
282 ax.set_title(titles[i],c=c,fontsize=fontsize,pad=5)
283
284 else:
285 ax.annotate(string.ascii_lowercase[i], xy=(-0.25, 1), xycoords='axes fraction',
286 xytext=(0, 0), textcoords='offset points', va='top', c=axcol,
287 fontsize=fontsize)
288
289
290
291 ax.axis('off')
292 # axins.axis('off')
293 axins.set_xticks([])
294 axins.set_yticks([])
295 axins.set_facecolor([0]*4)
296
297
298 for spine in axins.spines.values():
299 spine.set_color(color)
300 spine.set_linewidth(lwa)
301
302
303# plt.subplots_adjust(wspace=-0,hspace=0)
304plt.subplots_adjust(wspace=-.35,hspace=.5)
305
306
307# Display the plot
308plt.show()
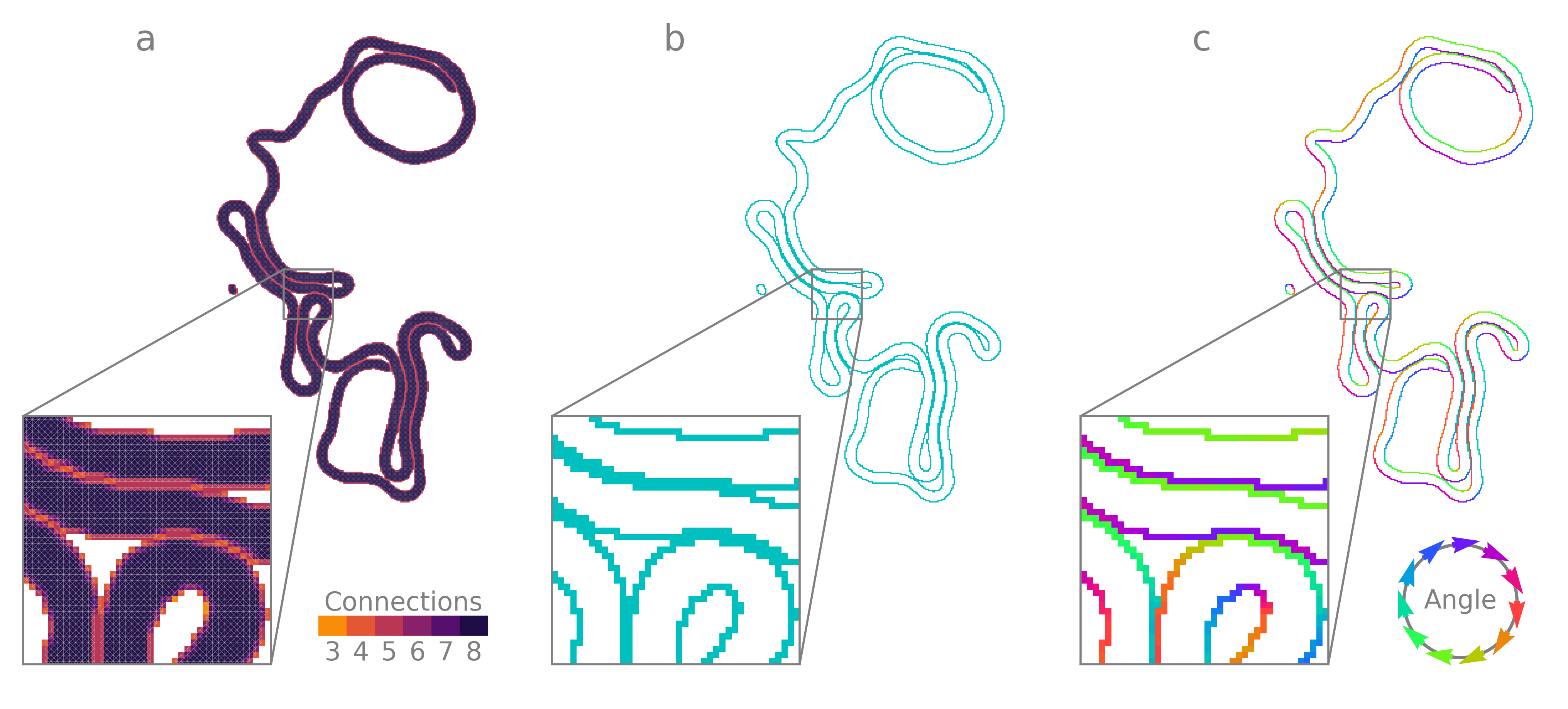
Pixels (or in ND, hypervoxels) may be classified as interior or boundary by their net connectivity. An ND hypervoxel connected to all \(3^{N}-1\) neighbors is classified as internal (8 in 2D, fully 2-connected to both cardinal and ordinal neighbors). Hypervoxels with fewer than \(3^{N}-1\) connections are classified as boundary. In Omnipose, hypervoxels with fewer than \(N\) connections are pruned when using affinity segmentation to avoid spurs and allow cell contours in 2D to be traced.
Because connections in an affinity graph are symmetrical, interfaces between objects are 2 hypervoxels thick. That is, the shortest path between the interiors of any two objects will pass through at least two boundary hypervoxels, one belonging to each object. Thresholding-based methods of boundary detection do not guarantee this symmetry and thus predict too many or too few boundary hypervoxels.