Cell diameter#
The idea of an average cell diameter sounds intuitive, but the standard implementation of this idea fails to capture that intuition. The go-to method (adopted in Cellpose) is to calculate the cell diameter as the diameter of the circle of equivalent area. As I will demonstrate, this fails for anisotropic (non-circular) cells. As an alternative, I devised the following simple diameter metric:
diameter = 2*(dimension+1)*np.mean(distance_field)
Because the distance field represents the distance to the closest boundary point, it naturally captures the intrinsic 'thickness' of a region (in any dimension). Averaging the field over the region (the first moment of the distribution) distills this information into a number that is intuitively proportional to the thickness of the region. For example, if a region is made up of a bungle of many thin fragments, its mean distance is far smaller than the mean distance of the circle of equivalent area. But to call it a 'diameter', I wanted this metric to match the diameter of a sphere in any dimension. So, by calculating the average of distance field of an n-sphere, we get the above expression for the the diameter of an n-sphere given the average of the distance field over the volume.
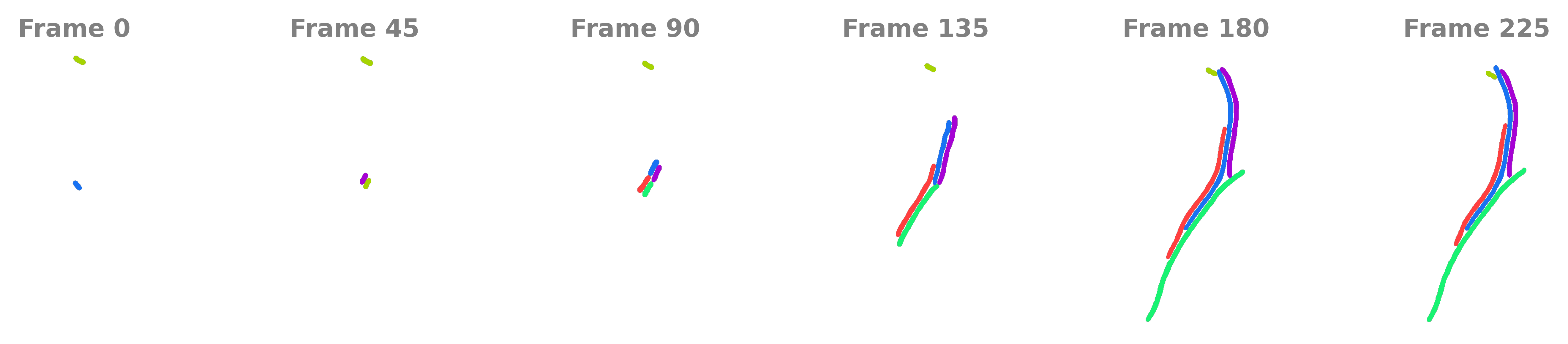
Example cells#
Filamenting bacterial cells often exhibit constant width but increasing length. This dataset comes from the deletion of the essential gene ftsN in Acinetobacter baylyi.
Show code cell source
1from pathlib import Path
2from cellpose_omni import utils, plot, models, io, dynamics
3import os, sys, io
4import numpy as np
5import matplotlib.pyplot as plt
6plt.style.use('dark_background')
7import matplotlib as mpl
8%matplotlib inline
9mpl.rcParams['figure.dpi'] = 600
10
11# Save a reference to the original stdout stream
12old_stdout = sys.stdout
13
14# Redirect stdout to a StringIO object
15sys.stdout = io.StringIO()
16
17
18import omnipose
19from omnipose.plot import imshow
20import tifffile
21omnidir = Path(omnipose.__file__).parent.parent
22basedir = os.path.join(omnidir,'docs','_static')
23nm = 'ftsZ'
24masks = tifffile.imread(os.path.join(basedir,nm+'_masks.tif'))
25mnc = omnipose.plot.apply_ncolor(masks)
26
27f = 1
28c = [0.5]*3
29fontsize=10
30dpi = mpl.rcParams['figure.dpi']
31Y,X = masks.shape[-2:]
32szX = max(X//dpi,2)*f
33szY = max(Y//dpi,2)*f
34
35# T = [50,80,100,150,180,240]
36T = range(0,len(masks),45)
37titles = ['Frame {}'.format(t) for t in T]
38ims = [mnc[t] for t in T]
39N = len(titles)
40
41fig, axes = plt.subplots(1,N, figsize=(szX*N,szY))
42fig.patch.set_facecolor([0]*4)
43
44for i,ax in enumerate(axes):
45 ax.imshow(ims[i])
46 ax.axis('off')
47 ax.set_title(titles[i],c=c,fontsize=fontsize,fontweight="bold")
48
49plt.subplots_adjust(wspace=0.1)
50plt.show()
51
52# Restore the original stdout stream
53sys.stdout = old_stdout
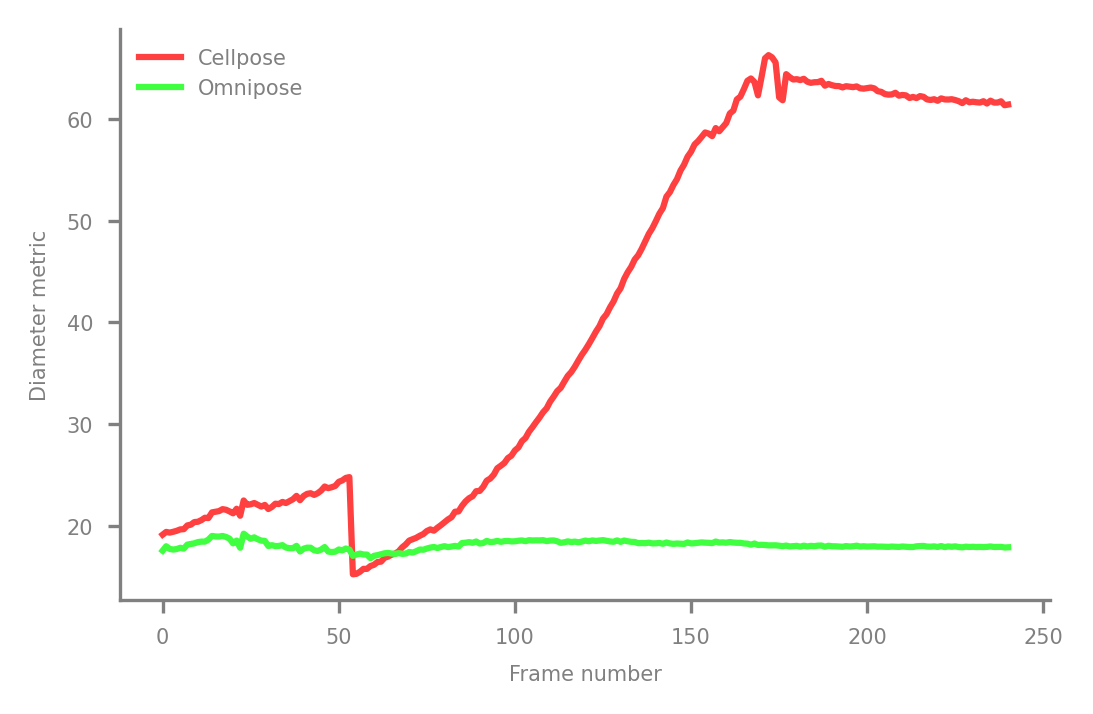
Compare diameter metrics#
By plotting the mean diameter (averaged over all cells after being computed per-cell, of course), we find that
the 'circle diameter metric' used in Cellpose rises drastically with cell length, but the 'distance diameter metric' of Omnipose remains nearly constant. If we tried to use the former to train a SizeModel(), images would get downsampled
heavily to the point of cells being too thin to segment, and that is assuming that the model can reliably detect the highly nonlocal property of cell length in an image instead of the local property of
cell width (at least, what we humans would point to and call cell width).
Show code cell source
1import fastremap
2n = len(masks)
3diam_old = []
4diam_new = []
5cell_num = []
6x = range(n)
7for k in x:
8 m = masks[k]
9 fastremap.renumber(m,in_place=True)
10 cell_num.append(m.max())
11 diam_old.append(utils.diameters(m,omni=False)[0])
12 diam_new.append(utils.diameters(m,omni=True)[0])
13
14
15from omnipose.utils import sinebow
16golden = (1 + 5 ** 0.5) / 2
17sz = 4
18labelsize = 5
19
20%matplotlib inline
21
22plt.style.use('dark_background')
23mpl.rcParams['figure.dpi'] = 300
24
25axcol = [0.5]*3+[1]
26N = 3
27colors = sinebow(N,offset=0)
28background_color = [0]*4
29
30fig = plt.figure(figsize=(sz, sz/golden),frameon=False)
31fig.patch.set_facecolor(None)
32
33ax = plt.axes()
34
35ax.plot(range(n),diam_old,c=colors[1],label='Cellpose')
36ax.plot(range(n),diam_new,c=colors[N],label='Omnipose')
37
38ax.legend(loc='best', frameon=False,labelcolor=axcol, fontsize = labelsize)
39ax.tick_params(axis='both', which='major', labelsize=labelsize,length=3, direction="out",colors=axcol,bottom=True,left=True)
40ax.tick_params(axis='both', which='minor', labelsize=labelsize,length=3, direction="out",colors=axcol,bottom=True,left=True)
41ax.set_ylabel('Diameter metric', fontsize = labelsize,c=axcol)
42ax.set_xlabel('Frame number', fontsize = labelsize, c=axcol)
43ax.set_facecolor(background_color)
44
45for spine in ax.spines.values():
46 spine.set_color(axcol)
47
48ax.spines['top'].set_visible(False)
49ax.spines['right'].set_visible(False)
50
51plt.show()